Following in the last document, now what I want to do is to load the preprocessed BAM files into R session, a recommended software is MethylKit. After a set of actions, the final bam file for each sample looks like below. The folder is in RRBS-pipeline/1.Preprocess/NUDUP/Ready:
RRBS-pipeline/1.Preprocess/NUDUP/Ready$ ls
Bc-01_S1.sorted.dedup.bam Bc-03_S3.sorted.dedup.bam Bc-05_S5.sorted.dedup.bam
Bc-02_S2.sorted.dedup.bam Bc-04_S4.sorted.dedup.bam Bc-06_S6.sorted.dedup.bam1. Process Bismark Alignment Result
According to methylkit package manual, the first step we should do it process Bismark results, since Bismark only do alignment work, so the function processBismarkAln is designed to call CpG's methylation value from these bam file. It would return the results of each sample in a format as XXX_cpg.txt format. Below is a script is written do apply this function.
# Script to run processBismarkAln
library("methylKit")
directory <- "../1.Preprocess/NUDUP/Ready/"
files <- dir(directory)
# This line is only used to extract Sample Name.
file.name <- as.list(unname(sapply(files,function(x) strsplit(x,split="_")[[1]][1])))
files <- as.list(paste(directory,files,sep=""))
message("Processing Bismark Alignment...")
my.methRaw=processBismarkAln(location=files,
sample.id=file.name,
assembly="danRer11",
save.context=NULL,
read.context="CpG",
mincov=10,
minqual=20,
treatment=c(0,0,0,1,1,1),
save.folder="./")Above script would take some time, roughly 1 hour for 6 samples. Then for each sample, a cpg.txt file would be generated. I remember GemBS software also would generate this kind of file, which means if I use GemBS file, I can directly input it into Methylkit for future analysis. Also, in above script, I applied some parameter, like mincov=10, and minqual=20. These are default settings from the software manual.
These cpg.txt loos like below:
Bc-01_CpG.txt
chrBase chr base strand coverage freqC freqT
1.1110 1 1110 F 40 97.50 2.50
1.1167 1 1167 R 78 87.18 12.82
1.11585 1 11585 F 12 0.00 100.00
1.11603 1 11603 F 12 0.00 100.00
1.11621 1 11621 F 12 0.00 100.002. Read CpG
After CpG calling, then we can try read them all into R session, to form an matrix for downstream analysis. This part of code is:
message("Reading CpG into R with methylKit...")
CpGFiles <- as.list(paste("./",unlist(file.name),"_CpG.txt",sep=""))
sampleName <- c("WildType_1", "WildType_2", "WildType_3", "Mutation_1", "Mutation_2", "Mutation_3")
myobj <- methRead(CpGFiles,
sample.id= as.list(sampleName),
assembly="danRer11",
treatment=c(0,0,0,1,1,1),
context="CpG")One amazing part for MethylKit is that we don't need to prepare reference genome ourselves. In above code, the sampleName is important. And treatment parameter indicates case/control status, which is quite important and will be used across the whole project.
3. Density Plot and Coverage Plot
This step is similar to QC plot check in methylation array, here I plotted the density histogram for these samples. MethylKit provided a function to do so:
par(mfrow=c(2,3))
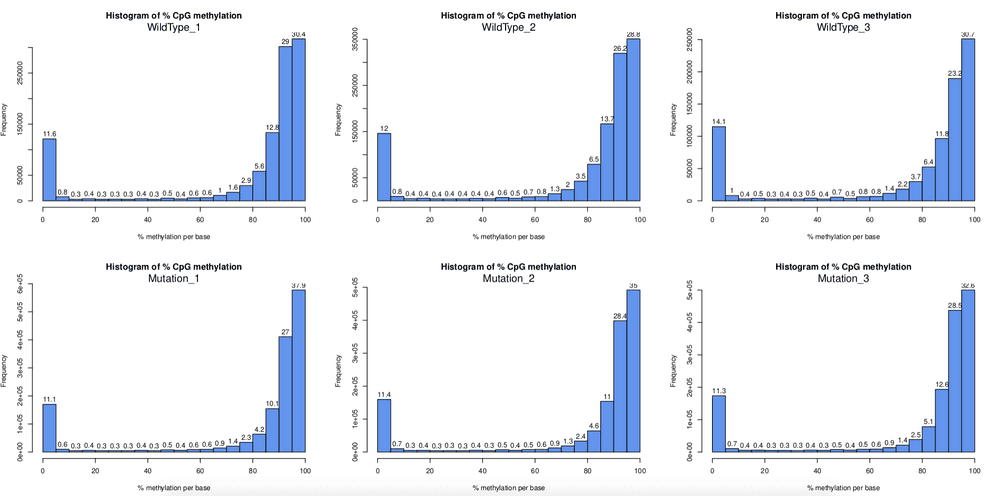
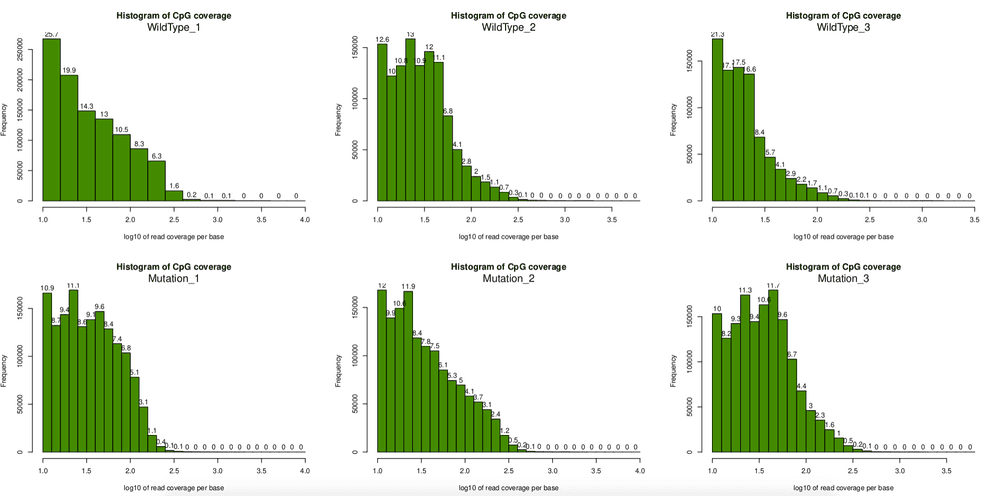
for(i in 1:length(myobj)) getMethylationStats(myobj[[i]],plot=TRUE,both.strands=FALSE)According to above plot, it seems the density plot looks very good, it shows the two peak I have been looking for. This is possiblly the most convinient function to check density plot for RRBS/WGBS data. Simiarly, I can get coverage plot:
4. Coverage Filtering and Normalisation
This step is important, it will filter CpGs with too low coverage (below 10), or redicularsly high (based on a null distribution). And normalisation would be apply afterward to across all samples to removed bias. The code is below:
message("Filtering Coverage")
filtered.myobj <- filterByCoverage(myobj,lo.count=10,lo.perc=NULL, hi.count=NULL,hi.perc=99.9)
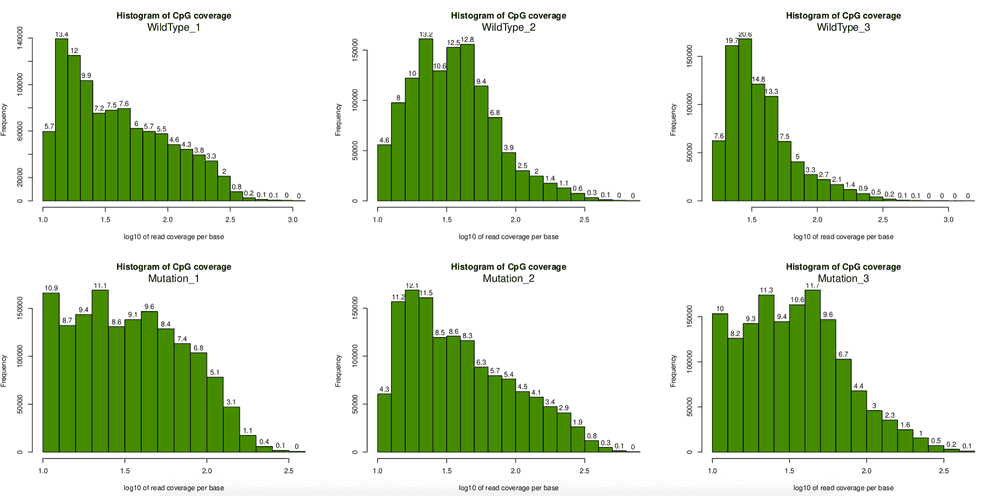
filtered.normed.myobj <- normalizeCoverage(filtered.myobj)After filtering, I tried to plot the coverage again, clearly some very high coverage bases are removed. This step is important, otherwise the comparision analysis later would be hugely biased.
5. Create Meth Object
Then finally we can create a meth object, this is the first human readable object we can create from methylKit package. The code is simply:
> meth <- unite(filtered.myobj, destrand=FALSE)
> meth
methylBase object with 554917 rows
--------------
chr start end strand coverage1 numCs1 numTs1 coverage2 numCs2 numTs2
1 1 1110 1110 + 46 45 1 55 45 10
2 1 1167 1167 - 89 78 11 35 33 2
3 1 12008 12008 + 197 1 196 86 1 85
4 1 12023 12023 + 198 0 198 86 0 86
5 1 12038 12038 + 198 0 198 83 0 83
6 1 12039 12039 - 218 0 218 104 0 104
...Also meth object contains the number of total CpGs we get across all samples:
> dim(meth)
[1] 554917 22
>A very important step is here, to generate beta matrix, this matrix may not help for DMR/DMP calling, but it's definitly useful for visualisation. A beta matrix here could allow me to implement a lot of array based algorithm on to sequence based data.
> beta <- percMethylation(meth)/100
> dim(beta)
[1] 554917 6
> head(beta)
WildType_1 WildType_2 WildType_3 Mutation_1 Mutation_2 Mutation_3
[1,] 0.978260870 0.81818182 0.8333333 0.9302326 0.907216495 0.846153846
[2,] 0.876404494 0.94285714 0.9428571 0.9056604 0.862385321 0.830188679
[3,] 0.005076142 0.01162791 0.0000000 0.0000000 0.003649635 0.005263158
[4,] 0.000000000 0.00000000 0.0000000 0.0000000 0.007272727 0.005263158
[5,] 0.000000000 0.00000000 0.0000000 0.0000000 0.003663004 0.000000000
[6,] 0.000000000 0.00000000 0.0000000 0.0000000 0.006211180 0.000000000
>So above are two very important file generate from methylKit, meth and beta. A lot of downstream analysis would be done based on them.